3DGS Diet
deepdaiv
Memory optimization for 3D Gaussian Splatting
Project Page
Detailed information about the project can be found in the project page above!
Project Overview
3D Gaussian Splatting memory optimization project was conducted.
DBSCAN Clustering was applied to reduce the number of Gaussians.
Training progress was monitored using wandb, and rendering results were visualized.
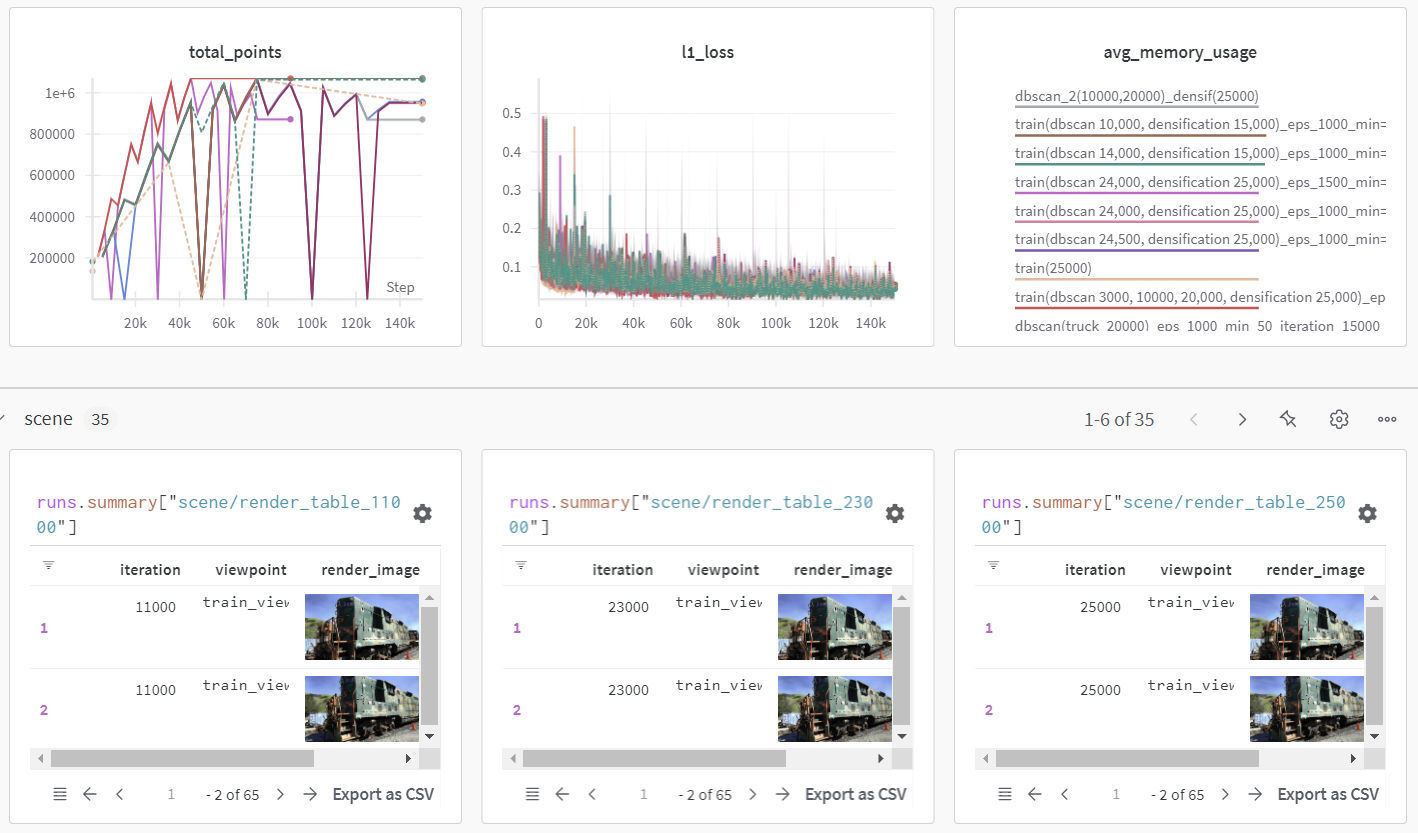
Result
The experiment results show that rendering performance (SSIM, PSNR, LPIPS) remains largely unchanged, while the number of Gaussians decreased by approximately 200,000.
| Model Application Point | Densification Stop Point | SSIM | PSNR | LPIPS | # Gaussian |
|---|---|---|---|---|---|
| 3DGS | 15,000 | 0.8756 | 24.449 | 0.1506 | 1,072,083 |
| DBSCAN_14000 | 15,000 | 0.8762 | 24.629 | 0.1500 | 1,064,677 |
| DBSCAN_24000 | 25,000 | 0.8651 | 24.081 | 0.1626 | 867,287 |
Rendering Visualization
Clustering applied at: 24,000 / Densification stopped at: 25,000

After conducting this project, we further experimented and wrote the paper Developing a Model for Improving 3D Gaussian Splatting Performance Based on DBSCAN.